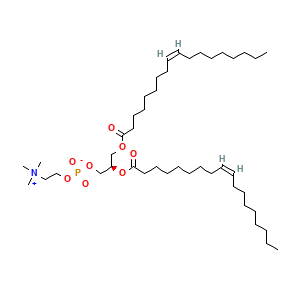
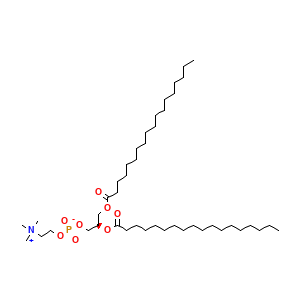
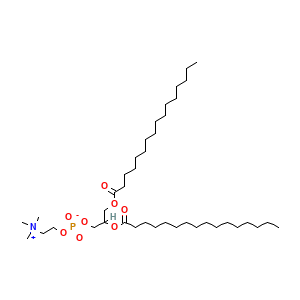
Complex Structure Info:
PDB ID:6VMS
Complex Structure type:
Complex Structure (proteins and ligands)
Interaction Data:
Complex ID:
8004
Protein(s) involved:
Protein ID:
Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
Molecules used in this complex structure:
Dynamic(s) in which this complex structure appears:
Dynamics ID: 1242
References in which this complex structure is mentioned:
Marie-Lise Jobin, Véronique De Smedt-Peyrusse, Fabien Ducrocq, Rim Baccouch, Asma Oummadi, Maria Hauge Pedersen, Brian Medel-Lacruz, Maria-Florencia Angelo, Sandrine Villette, Pierre Van Delft, Laetitia Fouillen, Sébastien Mongrand, Jana Selent, Tarson Tolentino-Cortez, Gabriel Barreda-Gómez, Stéphane Grégoire, Elodie Masson, Thierry Durroux, Jonathan A. Javitch, Ramon Guixà-González, Isabel D. Alves, Pierre Trifilieff. 2023. Impact of membrane lipid polyunsaturation on dopamine D2 receptor ligand binding and signaling. Molecular Psychiatry (). doi: 10.1038/s41380-022-01928-6.