Simulation report
Technical information
General information
Name: Delta opioid receptor (human)
PDB id: 4N6H
Activation state: Inactive
Description: Classical unbiased (NVT ensemble) complex flexibility assay.
Submitted by: GPCR drug discovery group (Pompeu Fabra University)
System setup
Solvent type: TIP3P
Membrane type: Homogeneous
Membrane composition: POPC
Ionic composition: Sodium ion (129 mM), Chloride (195 mM)
Number of molecules:
Water: 13363
POPC: 160
Chloride: 47
Sodium ion: 31
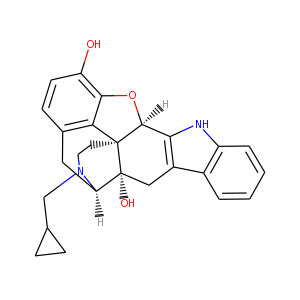
Naltrindole: 1
Delta-type opioid receptor: 1
Total number of atoms: 66513
Simulation details
Software and version: ACEMD, 3203
Forcefield and version: charmm, 36
Time step : 4.0 fs
Delta : 4.0 ns
Replicates: 1
Accumulated simulation time: 6.144 µs
Simulation components
Ligands
Receptor
Mutations
Table of simulated mutations
(Mutations shown in blue at the viewer)
| Residue ID | Residue seq. position | Generic num. | From | To |
|---|---|---|---|---|
| 72 | 37 | - | P | S |