Simulation report
Technical information
General information
Name: Adenosine receptor a2a in complex cgs-21680
PDB id: 4UHR.A
Activation state: Active
Description: Classical unbiased (NVT ensemble) complex flexibility assay.
Submitted by: GPCRmd community
System setup
Solvent type: TIP3P
Membrane type: Homogeneous
Membrane composition: POPC
Ionic composition: Sodium ion (157 mM), Chloride (191 mM)
Number of molecules:
Water: 18053
POPC: 192
Chloride: 62
Sodium ion: 51
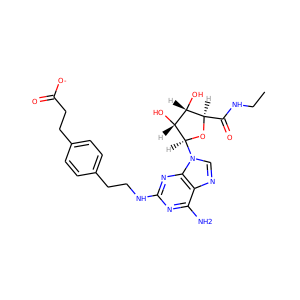
CGS-21680: 1
Adenosine receptor A2a: 1
Total number of atoms: 85036
Simulation details
Software and version: ACEMD, GPUGRID
Forcefield and version: CHARMM, 36m Feb 2016
Time step : 4.0 fs
Delta : 0.2 ns
Replicates: 3
Accumulated simulation time: 1.2 µs
Additonal parameters: Available at "Simulation protocol & starting files"
References
Ismael Rodríguez-Espigares, Mariona Torrens-Fontanals, et al.. 2020. GPCRmd uncovers the dynamics of the 3D-GPCRome. Nature methods 17 (8). doi: 10.1038/s41592-020-0884-y. (http://doi.org/10.1038/s41592-020-0884-y)
Find a list of all simulations published in this paper: 1469
Simulation components
Ligands
Kd: 632.0 nM More details
Kd: 4.4 nM More details
Kd: 4.4 nM More details
Kd: 4.4 nM More details
Kd: 7.6 nM More details
Kd: 27.0 nM More details
Kd: 7.6 nM More details
Kd: 74.0 nM More details
Kd: 4.3 nM More details
Kd: 12.0 nM More details
Kd: 99.0 nM More details
Kd: 36.0 nM More details
Kd: 22.0 nM More details
EC50: 18.0 nM More details
EC50: 18.0 nM More details
Receptor
Mutations
No mutations found