Simulation report
Technical information
General information
Name: Structure of beta2 adrenoceptor (apoform)
PDB id: 4LDO
Activation state: Active
Submitted by: Andrea Catte, University of Cagliari
System setup
Solvent type: TIP3P
Membrane type: Heterogeneous
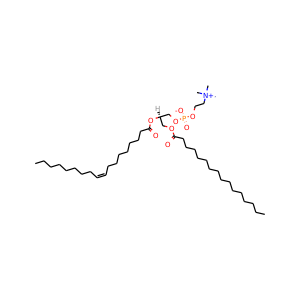
Membrane composition: POPC (80.0%), CHL1 (20.0%)
Ionic composition: Sodium ion (350 mM), Chloride (394 mM)
Number of molecules:
Water: 10000
POPC: 260
Chloride: 71
Cholesterol: 65
Sodium ion: 63
Levodopa: 1
Beta-2 adrenergic receptor: 1
Total number of atoms: 117654
Simulation details
Software and version: GROMACS, 2019.6
Forcefield and version: CHARMM, CHARMM36m
Time step : 2.0 fs
Delta : 0.5 ns
Replicates: 5
Accumulated simulation time: 1.0005 µs
Additonal parameters: Available at "Simulation protocol & starting files"
References
Biswas, AD; Catte, A; Mancini, G; Barone, V.. 2021. Analysis of L-DOPA and droxidopa binding to human β2-adrenergic receptor. Biophysical journal. doi: 10.1016/j.bpj.2021.11.007. (https://doi.org/10.1016/j.bpj.2021.11.007)
Find a list of all simulations published in this paper: 1476
Simulation components
Receptor
Mutations
Table of simulated mutations
(Mutations shown in blue at the viewer)
| Residue ID | Residue seq. position | Generic num. | From | To |
|---|---|---|---|---|
| 58 | 29 | 1x28 | D | X |