Simulation report
Technical information
General information
Name: Ffa2_tug1375_az1729_gi1-tug1375-az1729-gi1-mutateffa2-y117a
PDB id: 9CM7
Description: imulation studies the structural consequences of Y117A mutation by simulating FFA2 complex with orthosteric and allosteric agonists and G-protein. It produced NPT ensemble from 3 replicas of 1000 ns Langevin dynamics (T=310.15K, gamma=1ps-1) with semi-isotropic Berendsen barostat (P=1bar, tau=1ps) and 9A noncovalent cutoff.
Submitted by: Abdul-Akim Guseinov, University of Glasgow
System setup
Solvent type: TIP3P
Membrane type: Homogeneous
Number of molecules:
(2r,4r)-2-(2-chlorophenyl)-3-(4-(3,5-dimethylisoxazol-4-yl)benzoyl)thiazolidine-4-carboxylic acid: 1
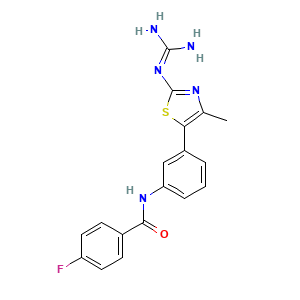
4-fluoro-n-[3-[2-[(aminoiminomethyl)amino]-4-methyl-5-thiazolyl]phenyl]benzamide: 1
Guanine nucleotide-binding protein G(i) subunit alpha-1: 1
Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1: 1
Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2: 1
Free fatty acid receptor 2: 1
Total number of atoms: 14524
Simulation details
Software and version: AMBER PMEMD.CUDA, 2020
Forcefield and version: ff19SB/lipid21/GAFF2, ff19SB/lipid21
Time step : 2.0 fs
Delta : 0.1 ns
Replicates: 3
Accumulated simulation time: 3.0 µs
Additonal parameters: Available at "Simulation protocol & starting files"
References
Xuan Zhang, Abdul-Akim Guseinov, Laura Jenkins, Alice Valentini, Sara Marsango, Katrine Schultz-Knudsen, Trond Ulven, Elisabeth Rexen Ulven, Irina G. Tikhonova, Graeme Milligan, Cheng Zhang. None. Allosteric modulation and biased signalling at free fatty acid receptor 2. None. doi: 10.1038/s41586-025-09186-6.
Find a list of all simulations published in this paper: 1561
Simulation components
Ligands
(2r,4r)-2-(2-chlorophenyl)-3-(4-(3,5-dimethylisoxazol-4-yl)benzoyl)thiazolidine-4-carboxylic acid
(1 molecule)

4-fluoro-n-[3-[2-[(aminoiminomethyl)amino]-4-methyl-5-thiazolyl]phenyl]benzamide
(1 molecule)
Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
(1 molecule)
Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
(1 molecule)
Receptor
Mutations
Table of simulated mutations
(Mutations shown in blue at the viewer)
2536
| Residue ID | Residue seq. position | Generic num. | From | To |
|---|---|---|---|---|
| 6 | 3 | - | C | X |
2537
| Residue ID | Residue seq. position | Generic num. | From | To |
|---|---|---|---|---|
| 2 | 1 | - | M | X |
2538
| Residue ID | Residue seq. position | Generic num. | From | To |
|---|---|---|---|---|
| 10 | 5 | - | N | X |
| 64 | 63 | - | L | X |
2535
| Residue ID | Residue seq. position | Generic num. | From | To |
|---|---|---|---|---|
| 117 | 117 | 34x53 | Y | A |
| 282 | 282 | 8x52 | A | - |
| 282 | 282 | 8x52 | F | - |
| 282 | 282 | 8x52 | G | - |
| 282 | 282 | 8x52 | R | - |
| 282 | 282 | 8x52 | G | - |
| 282 | 282 | 8x52 | L | - |
| 282 | 282 | 8x52 | Q | - |
| 282 | 282 | 8x52 | V | - |
| 282 | 282 | 8x52 | L | - |
| 282 | 282 | 8x52 | R | - |
| 282 | 282 | 8x52 | N | - |
| 282 | 282 | 8x52 | Q | - |
| 282 | 282 | 8x52 | G | - |
| 282 | 282 | 8x52 | S | - |
| 282 | 282 | 8x52 | S | - |
| 282 | 282 | 8x52 | L | - |
| 282 | 282 | 8x52 | L | - |
| 282 | 282 | 8x52 | G | - |
| 282 | 282 | 8x52 | R | - |
| 282 | 282 | 8x52 | R | - |
| 282 | 282 | 8x52 | G | - |
| 282 | 282 | 8x52 | K | - |
| 282 | 282 | 8x52 | D | - |
| 282 | 282 | 8x52 | T | - |
| 282 | 282 | 8x52 | A | - |
| 282 | 282 | 8x52 | E | - |
| 282 | 282 | 8x52 | G | - |
| 282 | 282 | 8x52 | T | - |
| 282 | 282 | 8x52 | N | - |
| 282 | 282 | 8x52 | E | - |
| 282 | 282 | 8x52 | D | - |
| 282 | 282 | 8x52 | R | - |
| 282 | 282 | 8x52 | G | - |
| 282 | 282 | 8x52 | V | - |
| 282 | 282 | 8x52 | G | - |
| 282 | 282 | 8x52 | Q | - |
| 282 | 282 | 8x52 | G | - |
| 282 | 282 | 8x52 | E | - |
| 282 | 282 | 8x52 | G | - |
| 282 | 282 | 8x52 | M | - |
| 282 | 282 | 8x52 | P | - |
| 282 | 282 | 8x52 | S | - |
| 282 | 282 | 8x52 | S | - |
| 282 | 282 | 8x52 | D | - |
| 282 | 282 | 8x52 | F | - |
| 282 | 282 | 8x52 | T | - |
| 282 | 282 | 8x52 | T | - |
| 284 | 283 | - | E | X |
Other molecules
(2r,4r)-2-(2-chlorophenyl)-3-(4-(3,5-dimethylisoxazol-4-yl)benzoyl)thiazolidine-4-carboxylic acid (allosteric lig.)
Specific State ID: 5402(1 molecule)

4-fluoro-n-[3-[2-[(aminoiminomethyl)amino]-4-methyl-5-thiazolyl]phenyl]benzamide (allosteric lig.)
Specific State ID: 5407(1 molecule)




