Simulation report
Technical information
General information
Name: Membrane-bound beta arrestin 1 impact of pip
PDB id: 6U1N
Activation state: Active
Description: -
Submitted by: Tomasz Stepniewski, IMIM
System setup
Solvent type: TIP3P
Membrane type: Heterogeneous
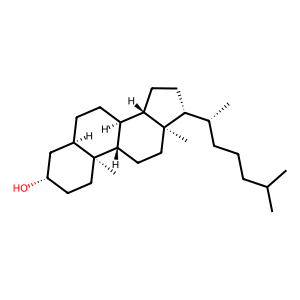
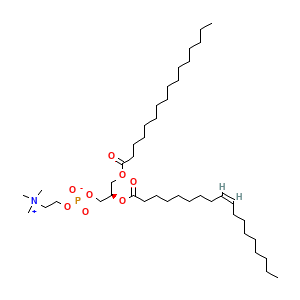
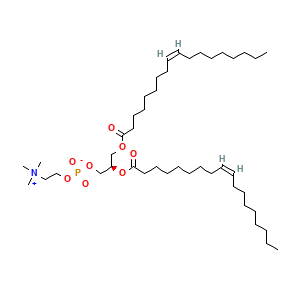
Membrane composition: CHL1 (10.0%), POPC (38.0%), DOPC (28.0%), DOPE (24.0%)
Ionic composition: Sodium ion (148 mM), Chloride (150 mM)
Number of molecules:
Water: 43994
POPC: 152
Chloride: 119
Sodium ion: 117
DOPC: 112
1,2-di-(9z-octadecenoyl)-sn-glycero-3-phosphoethanolamine: 96
Cholesterol: 40
Beta-arrestin-1: 1
Total number of atoms: 189204
Simulation details
Software and version: Acemd, 3.3
Forcefield and version: Charmm, 36M
Time step : 4.0 fs
Delta : 1.0 ns
Replicates: 1
Accumulated simulation time: 1.5 µs
Additonal parameters: Available at "Simulation protocol & starting files"
References
None. None. Location-biased β-arrestin conformations direct G protein-coupled receptor signa. None. doi: spatiotemporalsignaling.
Find a list of all simulations published in this paper: GPCRmd publication id 1526
None. None. Location-biased β-arrestin conformations direct G protein-coupled receptor signaling. None. doi: None.
Find a list of all simulations published in this paper: GPCRmd publication id 1527