Simulation report
Technical information
General information
Name: Fzd7 in inactive conformation (homo) (apoform)
PDB id: 9EPO
Description: -
Submitted by: Julien Bous, Karolinsk Institutet
System setup
Solvent type: TIP3P
Membrane type: Homogeneous
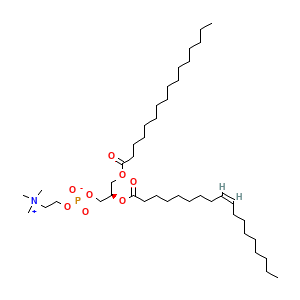
Membrane composition: Y01 (0.8%), POPC (99.2%)
Ionic composition: Chloride (194 mM), Sodium ion (145 mM)
Number of molecules:
Water: 13758
POPC: 129
Chloride: 48
Sodium ion: 36
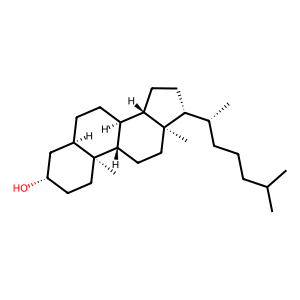
Cholesterol: 1
Frizzled-7: 1
Total number of atoms: 64511
Simulation details
Software and version: Gromacs, 2023-2
Forcefield and version: CHARMM, 36m
Time step : 2.0 fs
Delta : 0.1 ns
Replicates: 3
Accumulated simulation time: 1.8003000000000002 µs
Additonal parameters: Available at "Simulation protocol & starting files"
References
Bous, Julien and Kinsolving, Julia and Gratz, Lukas and Scharf, Magdalena M. and Voss, Jan Hendrik and Selcuk, Berkay and Adebali, Ogun and Schulte, Gunnar. 2024. Structural basis of frizzled 7 activation and allosteric regulation. Nature Communications 15 (1). doi: 10.1038/s41467-024-51664-4. (http://dx.doi.org/10.1038/s41467-024-51664-4)
GPCRmd publication page: 1528