Simulation report
Technical information
General information
Name: Tgr5 - system 4
PDB id: 7cfm
Activation state: Active
Description: -
Submitted by: M. Giovanna E. Papadopoulos, Philipps University Marburg
System setup
Solvent type: Implicit
Membrane type: Heterogeneous
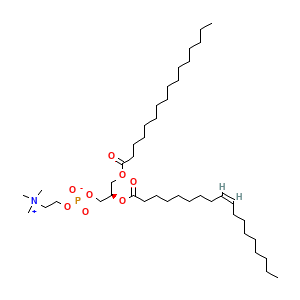
Membrane composition: POPC
Ionic composition: Sodium ion (150 mM), Chloride (176 mM)
Number of molecules:
Water: 25620
POPC: 212
Chloride: 81
Sodium ion: 69
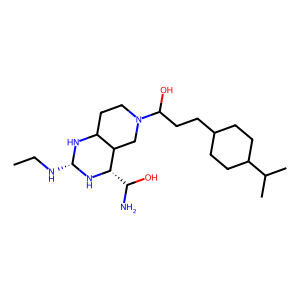
2-(ethylamino)-6-[3-(4-propan-2-ylphenyl)propanoyl]-7,8-dihydro-5h-pyrido[4,3-d]pyrimidine-4-carboxamide: 1
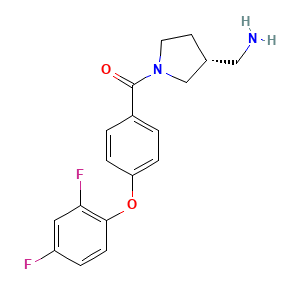
[(3r)-3-(aminomethyl)pyrrolidin-1-yl]-[4-(2,4-difluorophenoxy)phenyl]methanone: 1
G-protein coupled bile acid receptor 1: 1
Total number of atoms: 116581
Simulation details
Software and version: ACEMD, 3.2.3
Forcefield and version: CHARMM, 7.3.1
Time step : 4.0 fs
Delta : 1.0 ns
Replicates: 3
Accumulated simulation time: 1.5 µs
Additonal parameters: Available at "Simulation protocol & starting files"
References
Papadopoulos, M. Giovanna E. and Perhal, Alexander F. and Medel-Lacruz, Brian and Ladurner, Angela and Selent, Jana and Dirsch, Verena M. and Kolb, Peter. 2024. Discovery and characterization of small-molecule TGR5 ligands with agonistic activity. European Journal of Medicinal Chemistry. doi: 10.1016/j.ejmech.2024.116616. (http://dx.doi.org/10.1016/j.ejmech.2024.116616)
Find a list of all simulations published in this paper: GPCRmd publication id 1520
Simulation components
Ligands
(1 molecule)
[(3r)-3-(aminomethyl)pyrrolidin-1-yl]-[4-(2,4-difluorophenoxy)phenyl]methanone
(1 molecule)
Receptor
Mutations
Table of simulated mutations
(Mutations shown in blue at the viewer)
| Residue ID | Residue seq. position | Generic num. | From | To |
|---|---|---|---|---|
| 84 | 75 | 2x64 | W | A |
Other molecules
2-(ethylamino)-6-[3-(4-propan-2-ylphenyl)propanoyl]-7,8-dihydro-5h-pyrido[4,3-d]pyrimidine-4-carboxamide (allosteric lig.)
Specific State ID: 5348(1 molecule)

[(3r)-3-(aminomethyl)pyrrolidin-1-yl]-[4-(2,4-difluorophenoxy)phenyl]methanone (allosteric lig.)
Specific State ID: 5379(1 molecule)