Simulation report
Technical information
General information
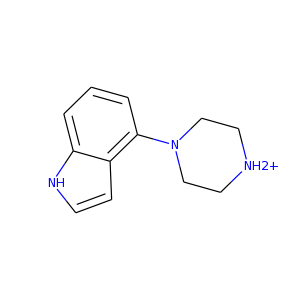
Name: Partially thermostabilized beta-1 adrenergic receptor in complex with 4-(1-piperazinyl)-1h-indole
PDB id: 3ZPQ.B
Activation state: Inactive
Description: Classical unbiased (NVT ensemble) complex flexibility assay.
Submitted by: GPCRmd community
System setup
Solvent type: TIP3P
Membrane type: Homogeneous
Membrane composition: POPC
Ionic composition: Chloride (207 mM), Sodium ion (164 mM)
Number of molecules:
Water: 16937
POPC: 189
Chloride: 63
Sodium ion: 50
4-(1-piperazinyl)-1h-indole: 1
Beta-1 adrenergic receptor: 1
Total number of atoms: 81132
Simulation details
Software and version: ACEMD, GPUGRID
Forcefield and version: CHARMM, 36m Feb 2016
Time step : 4.0 fs
Delta : 0.2 ns
Replicates: 3
Accumulated simulation time: 1.5 µs
Additonal parameters: Available at "Simulation protocol & starting files"
References
Ismael Rodríguez-Espigares, Mariona Torrens-Fontanals, et al.. 2020. GPCRmd uncovers the dynamics of the 3D-GPCRome. Nature methods 17 (8). doi: 10.1038/s41592-020-0884-y. (http://doi.org/10.1038/s41592-020-0884-y)
Find a list of all simulations published in this paper: 1469
Simulation components
Ligands
Receptor
Mutations
Table of simulated mutations
(Mutations shown in blue at the viewer)
| Residue ID | Residue seq. position | Generic num. | From | To |
|---|---|---|---|---|
| 314 | 282 | 6x27 | A | L |