Simulation report
Technical information
General information
Name: Atp-serca gamd rep 1 ext
PDB id: 3W5A
Description: 100 ns Gaussian accelerated Molecular Dynamics simulation of MG2+ATP-bound E1 SERCA in membrane. Force fields: FF19SB (protein), Lipid17 (membrane), OPC (water), GAFF2 (nucleotide). Final frame from 150 ns conventional MD is used as starting frame for GaMD simulation. Waters and membrane not included in uploaded files. This is an extension of previous 100 ns GaMD simulation.
Submitted by: Abigail Teitgen, University of California, San Diego
System setup
Solvent type: Other
Membrane type: Heterogeneous
Number of molecules:
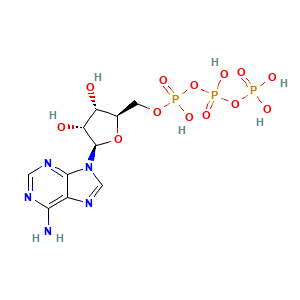
Adenosine-5'-triphosphate: 1
Magnesium: 1
Sarcoplasmic/endoplasmic reticulum calcium ATPase 2: 1
Total number of atoms: 15452
Simulation details
Software and version: Amber, 20
Forcefield and version: Amber, FF19SB
Time step : 2.0 fs
Delta : 0.02 ns
Replicates: 1
Accumulated simulation time: 0.1 µs
Additonal parameters: Available at "Simulation protocol & starting files"
References
Marcus T. Hock, Abigail E. Teitgen, Kimberly J. McCabe, Sophia P. Hirakis, Gary A. Huber, Michael Regnier, Rommie E. Amaro, J. Andrew McCammon, Andrew D. McCulloch. 2023. Multiscale Computational Modeling of the Effects of 2’-deoxy-ATP on Cardiac Muscle Calcium Handling. Journal of Applied Physics (134). doi: 10.1063/5.0157935.
Find a list of all simulations published in this paper: GPCRmd publication id 1504
Simulation components
Ligands
(1 molecule)
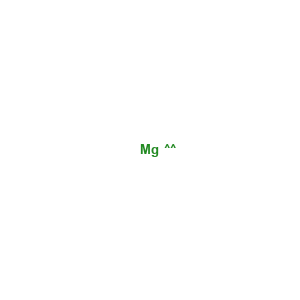
(1 molecule)
Sarcoplasmic/endoplasmic reticulum calcium ATPase 2
(1 molecule)
Receptor
Mutations
No mutations found



