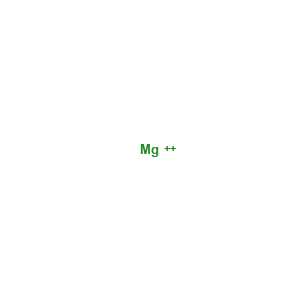Simulation report
Technical information
General information
Name: Ntsr1-gi (apoform)
PDB id: 6OS9
Activation state: Active
Submitted by: Nagarajan Vaidehi, Beckman Research Institute of the City of Hop
System setup
Solvent type: Implicit
Membrane type: Implicit
Number of molecules:
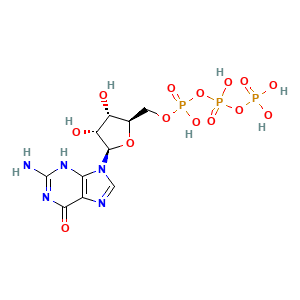
CID 6830: 1
Magnesium ion: 1
Guanine nucleotide-binding protein G(i) subunit alpha-1: 1
Total number of atoms: 16904
Simulation details
Software and version: GROMACS, 2022.1
Forcefield and version: CHARMM, 36m
Time step : 2.0 fs
Delta : 0.2 ns
Replicates: 1
Accumulated simulation time: 0.1602 µs
Additonal parameters: Available at "Simulation protocol & starting files"
Simulation components
Receptor
Mutations
Table of simulated mutations
| Residue ID | Residue seq. position | Generic num. | From | To |
|---|