Simulation report
Technical information
General information
Name: B2ar-icl3
PDB id: 6E67
Activation state: Active
Description: -
Submitted by: Nagarajan Vaidehi, Beckman Research Institute of the City of Hop
System setup
Solvent type: Implicit
Membrane type: Implicit
Number of molecules:
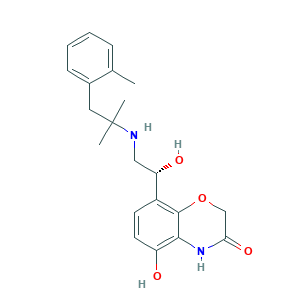
CHEMBL579394: 1
Beta-2 adrenergic receptor: 1
Total number of atoms: 5169
Simulation details
Software and version: GROMACS, 2021.3
Forcefield and version: CHARMM, 36m
Time step : 2.0 fs
Delta : 25.0 ns
Replicates: 1
Accumulated simulation time: 17.8 µs
Additonal parameters: Available at "Simulation protocol & starting files"
References
Sadler, Fredrik and Ma, Ning and Ritt, Michael and Sharma, Yatharth and Vaidehi, Nagarajan and Sivaramakrishnan, Sivaraj. 2023. Autoregulation of GPCR signalling through the third intracellular loop. Nature 615 (7953). doi: 10.1038/s41586-023-05789-z. (http://dx.doi.org/10.1038/s41586-023-05789-z)
GPCRmd publication page: 1507
Simulation components
Ligands
Receptor
Mutations
Table of simulated mutations
(Mutations shown in blue at the viewer)
| Residue ID | Residue seq. position | Generic num. | From | To |
|---|---|---|---|---|
| 82 | 82 | 2x66 | M | T |
| 84 | 84 | 23x49 | M | T |
| 173 | 173 | - | N | E |
| 213 | 212 | 5x65 | K | - |