Simulation report
Technical information
General information
Name: Or51e2 (apoform)
PDB id: 8F76
Activation state: Active
Submitted by: GPCRmd community
System setup
Solvent type: Implicit
Membrane type: Heterogeneous
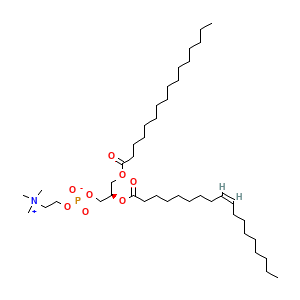
Membrane composition: POPC (87.3%), CHSD (12.7%)
Ionic composition: Chloride (147 mM), Potassium (205 mM)
Number of molecules:
Water: 16238
POPC: 165
Potassium: 60
Chloride: 43
Cholesterol hemisuccinate: 24
Olfactory receptor 51E2: 1
Total number of atoms: 77778
Simulation details
Software and version: GROMACS, 2021.3
Forcefield and version: CHARMM, 36m
Time step : 2.0 fs
Delta : 0.2 ns
Replicates: 1
Accumulated simulation time: 0.0002 µs
Additonal parameters: Available at "Simulation protocol & starting files"
References
Billesbølle, C.B., de March, C.A., van der Velden, W.J.C. et al.. 2023. Structural basis of odorant recognition by a human odorant receptor. Nature 615. doi: 10.1038/s41586-023-05798-y. (https://www.nature.com/articles/s41586-023-05798-y)
GPCRmd publication page: 1497
Simulation components
Receptor
Mutations
Table of simulated mutations
| Residue ID | Residue seq. position | Generic num. | From | To |
|---|