Simulation report
Technical information
General information
Name: B-arr2 wt
PDB id: 6U1N
Activation state: Active
Submitted by: GPCRmd community
System setup
Solvent type: TIP3P
Membrane type: Heterogeneous
Membrane composition: DOPE (24.0%), CHL1 (10.0%), POPC (38.0%), DOPC (28.0%)
Ionic composition: Sodium ion (149 mM), Chloride (167 mM)
Number of molecules:
Water: 36535
POPC: 152
DOPC: 112
Chloride: 110
Sodium ion: 98
Dope: 96
Cholesterol: 40
Beta-arrestin-2: 1
Total number of atoms: 166671
Simulation details
Software and version: Acemd, 3.5
Forcefield and version: Charmm, 36M
Time step : 4.0 fs
Delta : 1.0 ns
Replicates: 1
Accumulated simulation time: 1.0 µs
Additonal parameters: Available at "Simulation protocol & starting files"
References
Grimes, Jak and Koszegi, Zsombor and Lanoiselee, Yann and Miljus, Tamara and O'Brien, Shannon L. and Stepniewski, Tomasz M. and Medel-Lacruz, Brian and Baidya, Mithu and Makarova, Maria and Mistry, Ravi and Goulding, Joelle and Drube, Julia and Hoffmann, Carsten and Owen, Dylan M. and Shukla, Arun K. and Selent, Jana and Hill, Stephen J. and Calebiro, Davide. 2023. Plasma membrane preassociation drives β-arrestin coupling to receptors and activation. Cell 186 (10). doi: 10.1016/j.cell.2023.04.018. (http://dx.doi.org/10.1016/j.cell.2023.04.018)
GPCRmd publication page: 1493
Simulation components
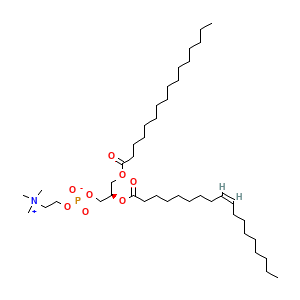
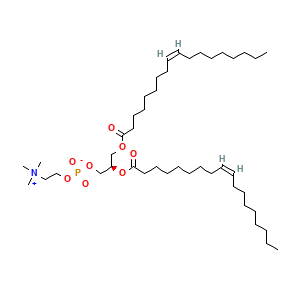
Ligands
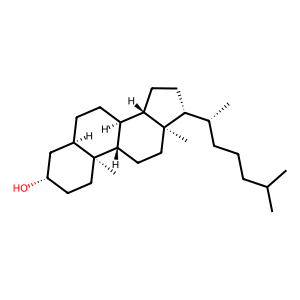
Receptor
Mutations
Table of simulated mutations
| Residue ID | Residue seq. position | Generic num. | From | To |
|---|