Simulation report
Technical information
General information
Name: Triple-mutant beta2 adrenergic receptor in complex with gs protein
PDB id: 3SN6
Activation state: Active
Submitted by: Nagarajan Vaidehi, Beckman Research Institute of the City of Hop
System setup
Solvent type: TIP3P
Membrane type: Homogeneous
Membrane composition: POPC
Ionic composition: Sodium ion (161 mM), Chloride (148 mM)
Number of molecules:
Water: 35234
POPC: 255
Sodium ion: 102
Chloride: 94
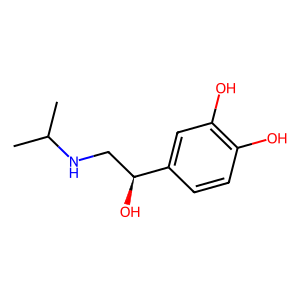
LEVISOPRENALINE: 1
Guanine nucleotide-binding protein G(s) subunit alpha isoforms short: 1
Total number of atoms: 156536
Simulation details
Software and version: GROMACS, 2019.3
Forcefield and version: CHARMM, 36m
Time step : 2.0 fs
Delta : 0.2 ns
Replicates: 1
Accumulated simulation time: 0.0002 µs
Additonal parameters: Available at "Simulation protocol & starting files"
References
Manbir Sandhu, Aaron Cho, Ning Ma, Elizaveta Mukhaleva, Yoon Namkung, Sangbae Lee, Soumadwip Ghosh, John H. Lee, David E. Gloriam, Stéphane A. Laporte, M. Madan Babu, Nagarajan Vaidehi. 2022. Dynamic spatiotemporal determinants modulate GPCR:G protein coupling selectivity and promiscuity. Nature Communications. doi: 10.1038/s41467-022-34055-5. (https://www.nature.com/articles/s41467-022-34055-5)
GPCRmd publication page: 1494
Simulation components
Ligands
Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
(1 molecule)
Receptor
Mutations
Table of simulated mutations
(Mutations shown in blue at the viewer)
| Residue ID | Residue seq. position | Generic num. | From | To |
|---|---|---|---|---|
| 212 | 174 | - | L | - |
| 212 | 174 | - | T | - |